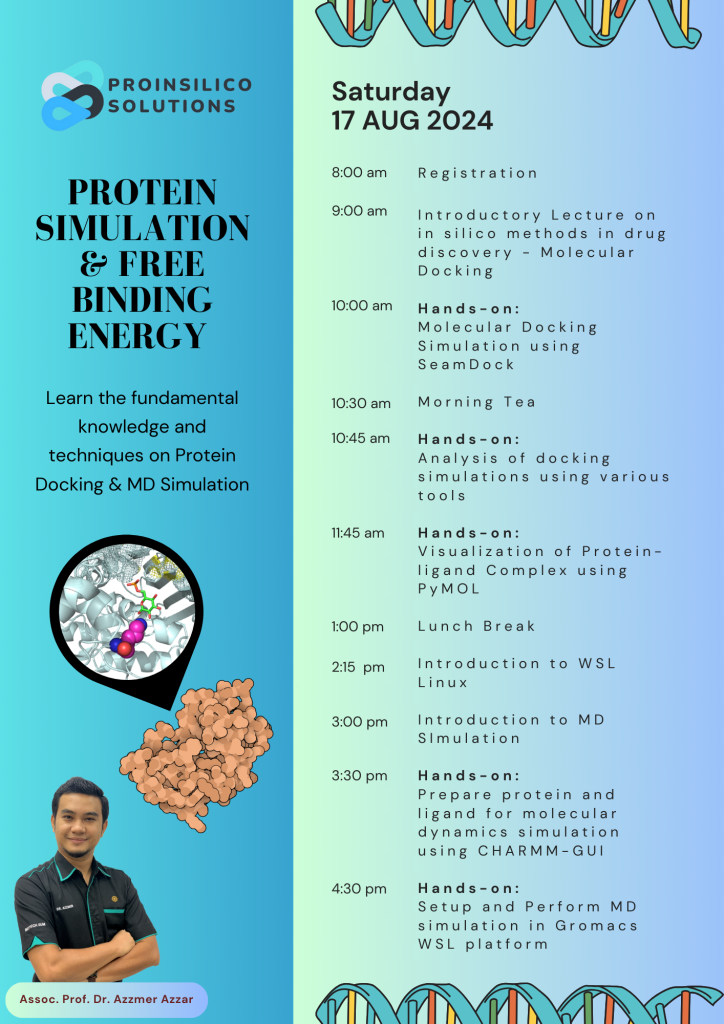
Workshop on Protein Simulation & Binding Free Energy using Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) Method

Calling all students, researchers, and scientists! Join our workshop led by experts in Structural Bioinformatics. Gain practical skills in biomolecular modeling, simulation, and data analysis with personalized guidance. Explore molecular docking and protein simulations on both Windows and Linux platforms, receiving expert support for software installation and technical assistance. Don’t miss this chance to enhance your skills—bring your own laptop!
In our workshop, you’ll master these skills:
• Conduct proteins and ligands docking using the SeamDock interactive webserver powered by Autodock Vina.
• Parameterize protein-ligand complexes with CHARMM_GUI.
• Set up and execute MD simulations using GROMACS software.
• Analyze protein-ligand interactions and process trajectories with Grace and VMD Software.
• Calculate Binding Free Energy using MMPBSA/MMGBSA techniques.
• Create illustrative images for publication using VMD and PyMol.
Join us now
https://lnkd.in/gyqJ-tiw




0 Comments